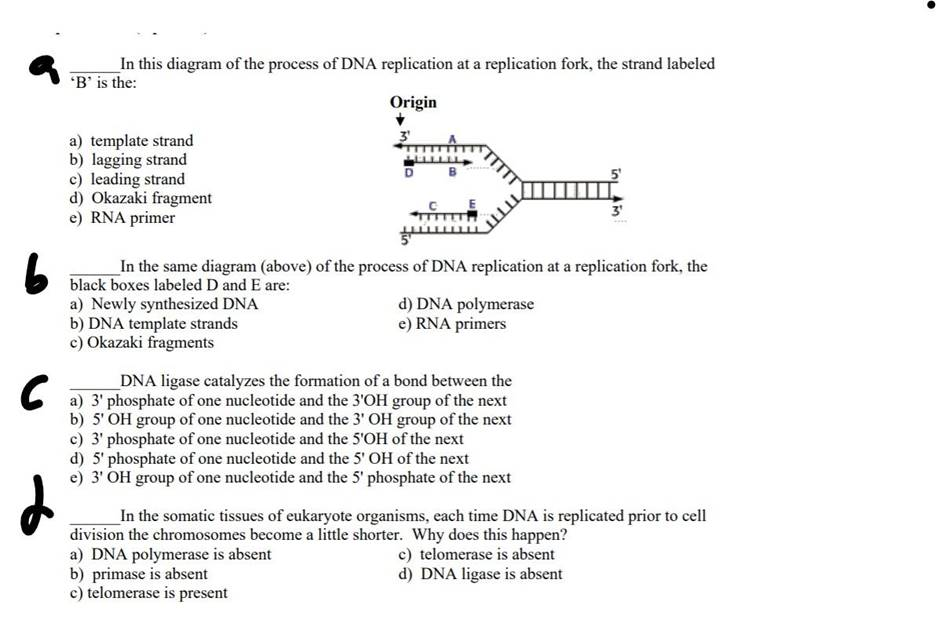
In this diagram of the process of DNA replication at a replication fork, the strand labeled ‘B’ is the: Origin D B a) template strand b) lagging strand c) leading strand d) Okazaki fragment e) RNA primer b In the same diagram (above) of the process of DNA replication at a replication fork, the black boxes labeled D and E are: a) Newly synthesized DNA d) DNA polymerase b) DNA template strands e) RNA primers c) Okazaki fragments ? 6 DNA ligase catalyzes the formation of a bond between the a) 3′ phosphate of one nucleotide and the 3’OH group of the next b) 5′ OH group of one nucleotide and the 3′ OH group of the next c) 3′ phosphate of one nucleotide and the 5’OH of the next d) 5′ phosphate of one nucleotide and the 5′ OH of the next e) 3′ OH group of one nucleotide and the 5′ phosphate of the next In the somatic tissues of eukaryote organisms, each time DNA is replicated prior to cell division the chromosomes become a little shorter. Why does this happen? a) DNA polymerase is absent c) telomerase is absent b) primase is absent d) DNA ligase is absent c) telomerase is present
The Correct Answer and Explanation is :
In the provided DNA replication fork diagram, the strand labeled ‘B’ is the lagging strand (option b). During DNA replication, the two parental DNA strands separate, forming a Y-shaped structure known as the replication fork. Each of these strands serves as a template for the synthesis of new complementary strands. The leading strand is synthesized continuously in the 5′ to 3′ direction, moving towards the replication fork. In contrast, the lagging strand (strand ‘B’) is synthesized discontinuously, away from the replication fork, in short segments called Okazaki fragments. This discontinuous synthesis occurs because DNA polymerase can only add nucleotides in the 5′ to 3′ direction, necessitating the repeated initiation of synthesis on the lagging strand as the replication fork progresses.
Regarding the black boxes labeled ‘D’ and ‘E’ in the same diagram, these represent RNA primers (option e). RNA primers are short sequences synthesized by the enzyme primase. They provide a starting point with a free 3′ hydroxyl group for DNA polymerase to begin adding DNA nucleotides. On the lagging strand, each Okazaki fragment starts with an RNA primer, which is later removed and replaced with DNA nucleotides, and the fragments are joined together by DNA ligase.
DNA ligase catalyzes the formation of a bond between the 3′ hydroxyl group of one nucleotide and the 5′ phosphate group of the next (option e). This enzyme is crucial for sealing nicks in the DNA backbone by forming phosphodiester bonds, thereby joining Okazaki fragments on the lagging strand and ensuring the continuity of the DNA molecule.
In eukaryotic somatic cells, chromosomes become slightly shorter with each round of DNA replication due to the absence of telomerase (option c). Telomerase is an enzyme that extends the telomeres, which are repetitive nucleotide sequences at the ends of chromosomes that protect them from degradation. Without sufficient telomerase activity, telomeres progressively shorten with each cell division, eventually leading to cellular senescence or apoptosis when they reach a critically short length.
Understanding these mechanisms is fundamental to molecular biology, as they explain how genetic information is accurately replicated and maintained, and how its dysregulation can lead to aging and disease.